|
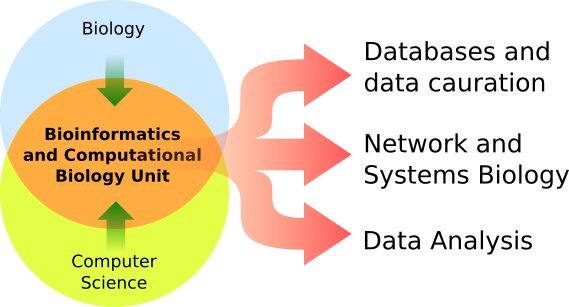
We work in close contact with experimental biologists to combine computational approaches with wet lab experiments. Our research is focused on Network and Systems Biology, Data Analysis, Data Curation and Storage. We process and analyze biological results by making algorithms and models. We performe statistical analysis on high-throughput data, Image Analysis and manual biological data curation continuing the MINT legacy with new services such as mentha and SIGNOR. Our philosophy is that bioinformatics should not be an end in itself, but rather be a bridge to connect classical biology with computational approaches. Our Lab is located at the University of Tor Vergata in Rome, Italy. (Leggi in italiano:  ) )
|
|||
|
Computational Facilities
The University of Rome Tor Vergata provides access to a 12MiBs Internet connection. Bioinformatics and Computational Biology Unit has more than 100TB of disk space used for data analysis and storage. The desktop computers and servers used by our unit are 5, excluding personal devices, replaced every 4-5 years. Computer scientists, software developers and data analysts work primarily on Unix/Linux environment. Our web services and resources are all hosted on Unix environment: MINT, mentha, SINGOR and others. MINT is our primary database and its curators work on Windows and Mac machines. We use Java, Tomcat, Jboss, Hibernate, R, PHP, Python and others, plus we archive our data in PostgreSQL and MySQL databases. Our main server has 16 cores and 24GB RAM. Our main computing machine has 12 cores and 24GB RAM. |
|||